Whiteley Lab Researchers Report Microbiome Controls How Well Pathogens Can Access Iron in Infections
Read the full article in PLOS Pathogens.
Iron is an essential nutrient for almost all of life - not only for us but also the bacterial pathogens that try to infect us. In the human body, free iron is actively kept in low amounts, and as a result bacterial growth is kept in check. This tactic of sequestering iron is one of the most vital ways that we fight infections, and in response, pathogens have evolved elaborate ways to plunder our body’s precious iron stores. However, most studies focus on how bacteria react to iron starvation in the lab – in plastic Petri dishes and glass test tubes – rather than in the vastly more complex environment that is our bodies. While these studies have laid important groundwork and have the advantage of being highly controlled, it is also necessary to not overlook the myriad factors that make up an actual host setting, including its resident microbiome. In fact, an intricate network of interactions between the pathogen interloper, the host, and the host’s microbiome likely dictates pathogen strategies for acquiring iron and establishing infection in the host.
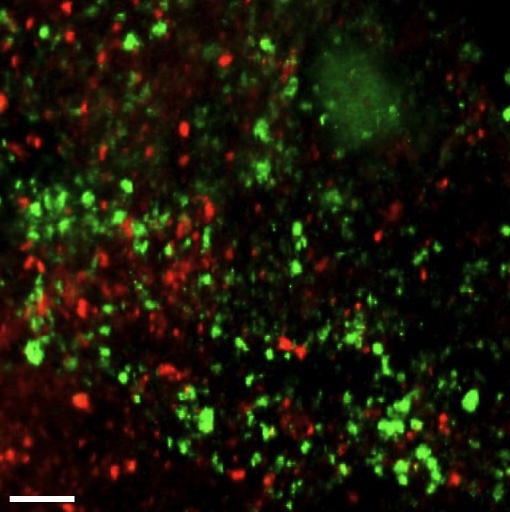
Figure 1: Confocal micrograph of a mouse skin abscess co-infected with the dental bacteria Aggregatibacter actinomycetemcomitans (Aa, red) and Streptococcus gordonii (Sg, green) (Visick et al., 2016). UT researchers discovered that in abscesses by itself the pathogen Aa is not restricted for iron, but upon introducing another bacterium, the health-associated Sg, Aa experiences iron limitation. Scale bar, 25 µm. Credit: Jake Everett and Kendra Rumbaugh of Texas Tech University Health Sciences Center.
Apollo Stacy and co-authors in the Whiteley Lab have reported in PLOS Pathogens that the bacterium Aggregatibacter actinomycetemcomitans (Aa) likely confronts very different levels of iron starvation in the host depending on the composition of its microbiome. Aa is a pathogen that lives in our mouths even when we are healthy, but sneakily can also cause what is known as aggressive periodontitis, a highly destructive gum disease that primarily affects adolescents and, if left untreated, can lead to rapid tooth loss. Unfortunately, Aa is not satisfied with just making our teeth fall out. It is also known to escape the mouth and form life-threatening abscesses in the brain and lungs.
In their new study, Whiteley’s team used next-generation sequencing to characterize how Aa gene expression changes when iron is removed from monoculture lab settings. They then looked at Aa gene expression in host mice infected with Aa alone or Aa along with another dental bacterium, the relatively harmless Streptococcus gordonii (Sg). Finally, they looked at Aa in human dental plaque, the slime that accumulates on our teeth when we don’t brush them. This slime is made up of a rich community of bacteria, not just Aa and Sg, and Whiteley’s team was able to inspect just Aa, specifically how its gene expression changes between paired healthy and diseased teeth from individual patients (Jorth et al., 2014). By comparing their monoculture lab data, where iron levels were known, to the ‘black box’ of the host, they could make conclusions about how iron availability toAa is affected by other bacteria in co-infections.
Whiteley’s sequencing studies centering on Aa gene expression under defined high and low iron levels in the lab was a useful starting strategy. That’s because, in response to low iron, bacteria are wired to increase expression of iron transporters and decrease expression of cellular processes that consume iron – a regulatory program that enables bacteria to go into ‘lockdown’ while greedily snatching up their favorite metal. By doing these initial lab studies, the researchers gained a better idea of how Aa deals with low iron. Using this dataset, they could then indirectly assess what iron levels are like in the host based on Aa gene expression in its primary infection sites, abscesses and the oral cavity.
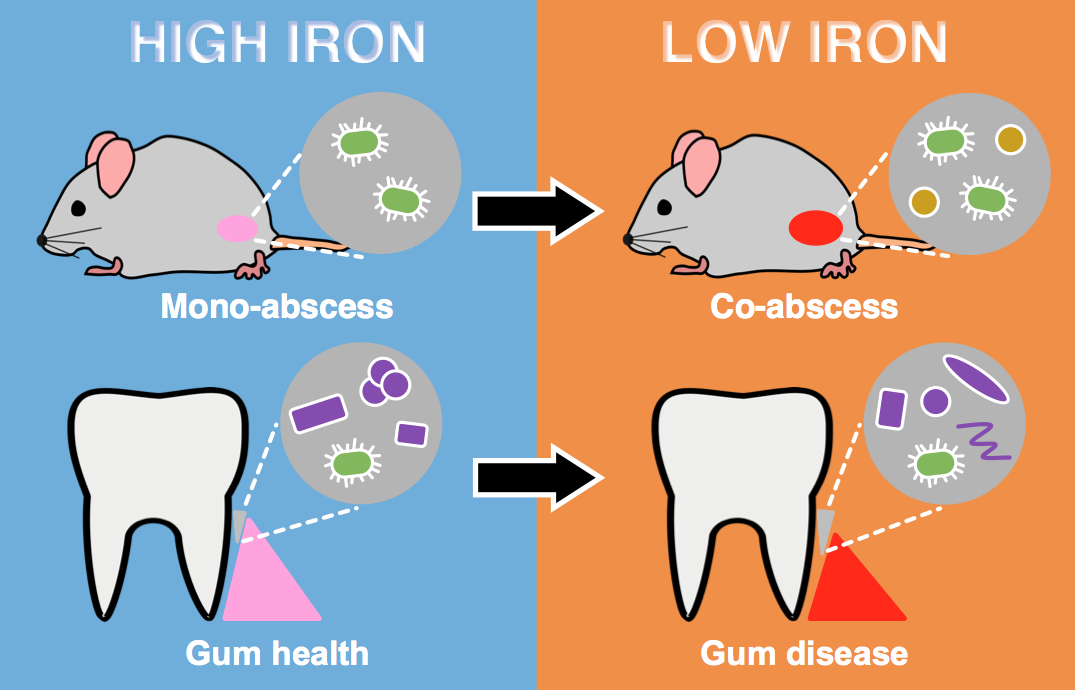
Figure 2: Model for how the microbial community impacts iron availability to the pathogen Aa. In mouse abscess mono-infection and healthy human dental plaque (purple cells), iron is more available to Aa (green cells). In mouse abscess co-infection with Sg (orange cells) and diseased human dental plaque (purple cells), iron is less available to Aa. The dental microbiome shifts in composition between health and disease.
These findings suggest that host iron availability is not only non-homogeneous but also dependent on the bacterial community. Future studies of human infections should therefore keep the microbiome in mind when designing therapies to target pathogens’ need for iron. When the bacterial community is just right, this Achilles’ heel of the pathogen could be especially exploitable (such as by temporarily keeping iron out of the diet).
Read the full article in PLOS Pathogens: Microbial Community Composition Impacts Pathogen Iron Availability During Polymicrobial Infection
References:
1. Visick et al., October 2016.