Overview
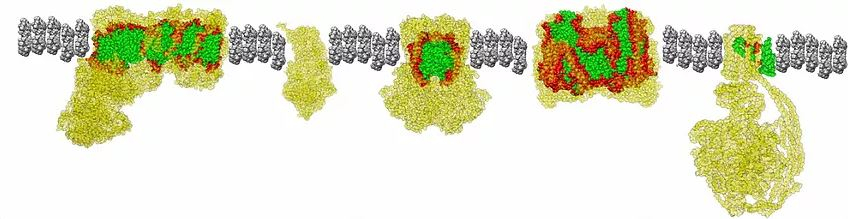
Most complex lifeforms - humans, animals, plants, and even unicelluar eukaryotes - are dependent on at least two genomes to generate their cellular energy: nuclear and mitochondrial. It is absolutely essential that the intricate interactions between gene products encoded by these two genomes remain in sync. Otherwise, organismal health and fitness decreases drastically.
Our research explores the evolution of such cytonuclear interactions, which has implications for a wide range of topics that are of general interest to biologists. These include the origin of the eukaryotes, endosymbiosis, the ultimate cause of sexual reproduction, why organisms age, how speciation occurs, and environmental adaptation, to name a few.
Ongoing projects -
Causes and consequences of mitochondrial mutations
Mutations in the mitochondrial genome underly multiple diseases and have been suggested to play a general role in aging. However, understanding the causes and consequences of mitochondrial mutations is limited by a focus on mammalian models, which uniformly high mitochondrial mutation rates. We are characterizing mitochondrial mutations and their effects on physiology in diverse eukaryotic systems, including invertebrates, plants, and micro-eukaryotes. We use high-fidelity sequencing to characterize rates and types of mitochondrial mutations across eukaryotes and under different environments (e.g., increased oxidative stress). Of particular interest is the frequency of C -> T transitions vs. G -> T transversions. The former are usually attributed to replication errors while the latter are caused by oxidative damage. Oxidative damage is implicated in aging theories, but replication errors have been shown to dominate the mutational landscape in mammalian mitochondrial genomes. We also use physiological techniques to quantify distinct states of oxidative phosphorylation, reactive oxygen species (ROS) production, and metabolic rate in systems with varying sources and rates of mitochondrial mutations. This research will further uncover the possibilities for mitochondrial mutations to influence cellular and organismal processes, with implications for human health, disease progression, and aging. This work is funded by an NIH Early Career MIRA award (1R35GM142836).
Some publications describing this work:
MtDNA mutations in C. elegans nematodes
MtDNA mutation spectra in PolG mice
Cytonuclear coevolution
Cytonuclear coevolution is expected to be common in eukaryotes to maintain proper cytonuclear interactions and function. Under cytonuclear coevolution, any changes in one genome must be complemented by appropriate changes in the other genome. We are using molecular data from a wide range of eukaryotes to test predictions of cytonuclear coevolution. We are also interested in what specific forms cytonuclear coevolution takes when it is found - one notable hypothesis is nuclear compensation, in which slightly deleterious mutations in cytoplasmic genomes are offset by compensatory changes in nuclear-encoded genes.
Some publications describing this work:
Causes and consequences of mito mutations
Sexually antagonistic mitonuclear coevolution
Mitonuclear evolutionary rate covariation in bivalves
Roles of cytoplasmic genomes during speciation
If mutations in cytoplasmic genomes cause the fixation of mutations in the nuclear genome, then cytoplasmic and nuclear genomes might be coadapted to one another within an evolutionary lineage. Hybridization between species or populations might break up these coadapted genomes, leading to hybrid breakdown and reproductive isolation between lineages. Paradoxically, cytoplasmic genomes are especially prone to cross species boundaries via introgression. To resolve these conflicting observations, we are using large, publicly available datasets and targeted experiments in swordtail fishes (in collaboration with Molly Schumer's lab) to reconcile the contradictory roles for cytoplasmic genomes at species boundaries.
Some publications describing this work:
The paradoxical roles of cytoplasmic genomes in speciation
A lethal mitonuclear incompatibility in swordtails
Mitochondrial function and environmental adaptation
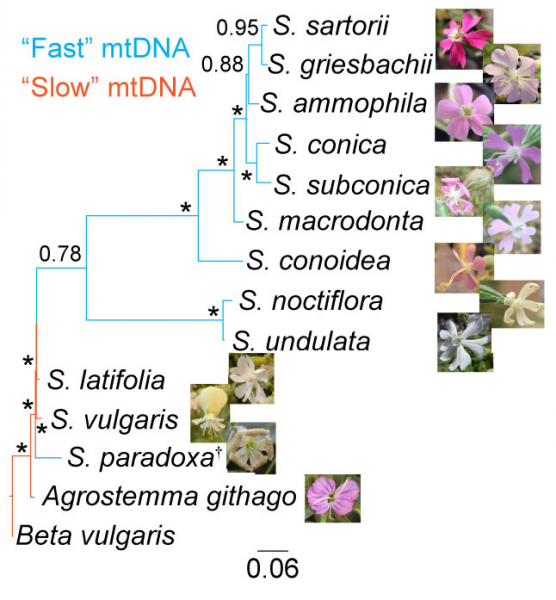
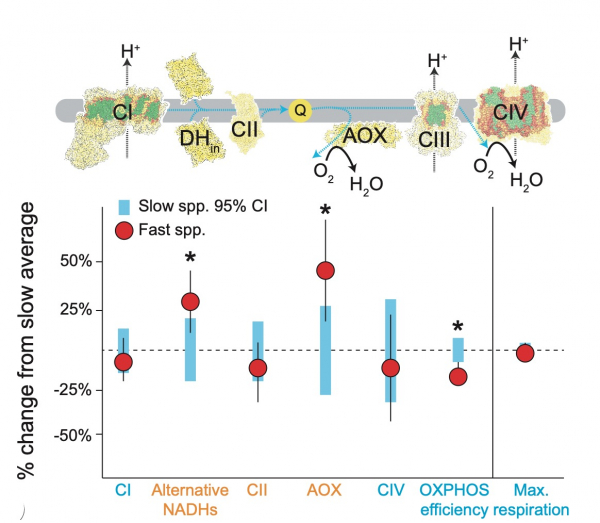
Because mitochondria provide the majority of cellular energy in eukaryotes, mitochondrial function is critical for adaptation to novel environments. We're doing detailed wet lab experiments looking at mitochondrial function in different systems and in different environments. For example, we previously examined how mitochondrial respiration and whole animal metabolism phenotypes may change in mountain aquatic insects during acclimation to different thermal environments. We also showed that Silene angiosperms with high mitochondrial mutation rates show increased reliance on nuclear-encoded components for mitochondrial function. We're exploring mitochondrial respiration, membrane potential, and ROS production in a variety of systems. We're also examining signatures of selection on mitochondrial genes from animals living across different environments.
Some publications describing this work:
Thermal responses of mitochondrial respiration in montane mayflies
Thermal responses of whole animal respiration across elevation and latitudes
How to measure thermal acclimation in biological rates
Differences in thermal responses across organizational levels
Silene angiosperms with different mt mutation rates have different mt physiology
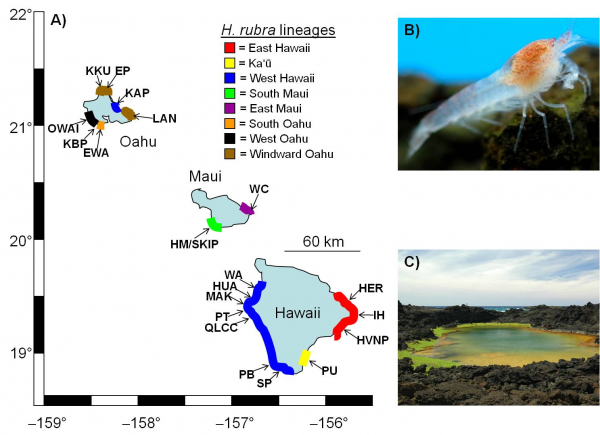
Anchialine ecology and evolution
Anchialine habitats and organisms are model systems in our work. Anchialine habitats are coastal water bodies influenced by both seawater and freshwater that have high levels of endemism. We are currently examining the physiological and genetic basis for red coloration in anchialine shrimps. These shrimps also have high rates of mitochondrial mutation and strong population structure across the islands where they're found. We also examine anchialine microbial ecology and animal-microbe interactions in anchialine habitats. New anchialine habitats were created in 2018 by lava flows in Hawaii and we're examining the formation and succession of their microbial communities (funded by an NSF RAPID award - NSF DEB2020099).
Some publications describing this work:
Carotenoid variation in red coloration across anchialine shrimp lineages
Microbial community structure in anchialine habitats
Seasonality in anchialine microbial communities
Microbial communities and vertical stratification in anchialine habitats
| havird_researchstrategy_norefs_arial_093020.pdf | 2.78 MB |