Research
Development and Applications of Photodissociation
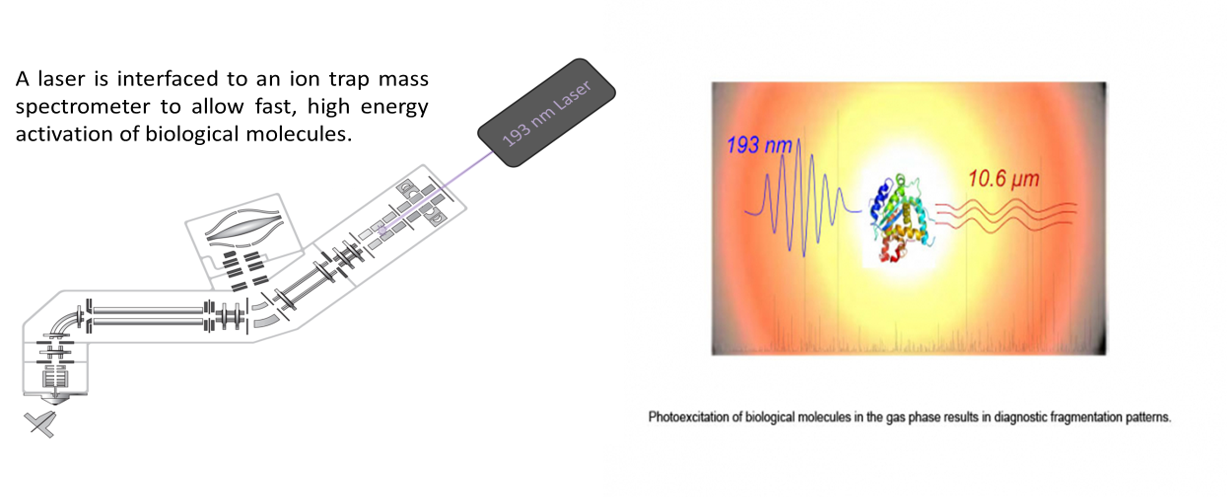
Despite the tremendous array of mass spectrometric tools which allow generation of large data sets, it has become clear that there remains a need for enhancing the selectivity of mass spectrometric strategies for problems involving complex mixtures or targeted screening while also offering versatility and tunability for other applications. Collision induced dissociation (CID) has the longest and richest history in the realm of ion activation methods, although its limited or poorly defined energy deposition has led to vigorous exploration of alternative strategies. Moreover, the sequence and charge state of biopolymers (like peptides, oligosaccharides, and oligonucleotides) can greatly influence dissociation. Newer techniques have shown promise both for widespread and niche applications, as well as offering complementary fragmentation patterns, more tunable energy deposition, or alternative dissociation mechanisms. We are developing IR and UV photodissociation (PD) methods and electron transfer dissociation (ETD), in conjunction with chemical derivatization methods that add chromophores and/or charge sites to molecules in order to modify ion fragmentation patterns. There are several key benefits of photodissociation that make a compelling case for continued development. For example, UVPD allows higher energy deposition (3 to 7 eV per photon depending on the wavelength) in a short time period (< 10 ns), allowing adaptation for fast, higher throughput applications. The ability to label molecules with IR or UV chromophores offers the ability to incorporate a high degree of selectivity into an MS/MS strategy. Our work is aimed at the structural characterization of biological molecules, especially proteins.
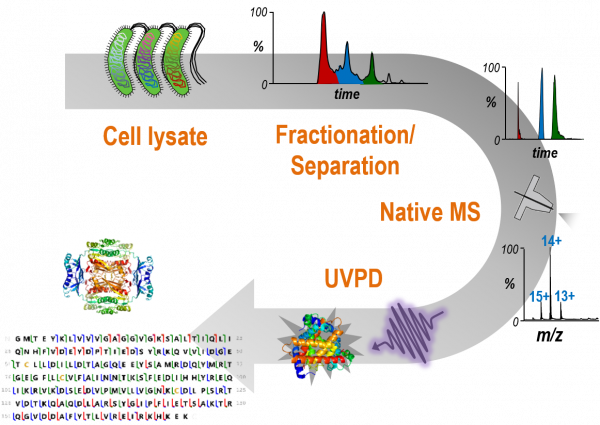
Characterization of Protein Complexes
We are developing methods for analysis of protein complexes based on native mass spectrometry. Intact protein complexes are fractionated and separated using capillary electrophoresis prior to dissection and characterization by ultraviolet photodissociation. Sequence maps are generated for the proteins in the complexes based on the fragmentation patterns, and conformations of proteins are characterized by measurements of collision cross-sections.
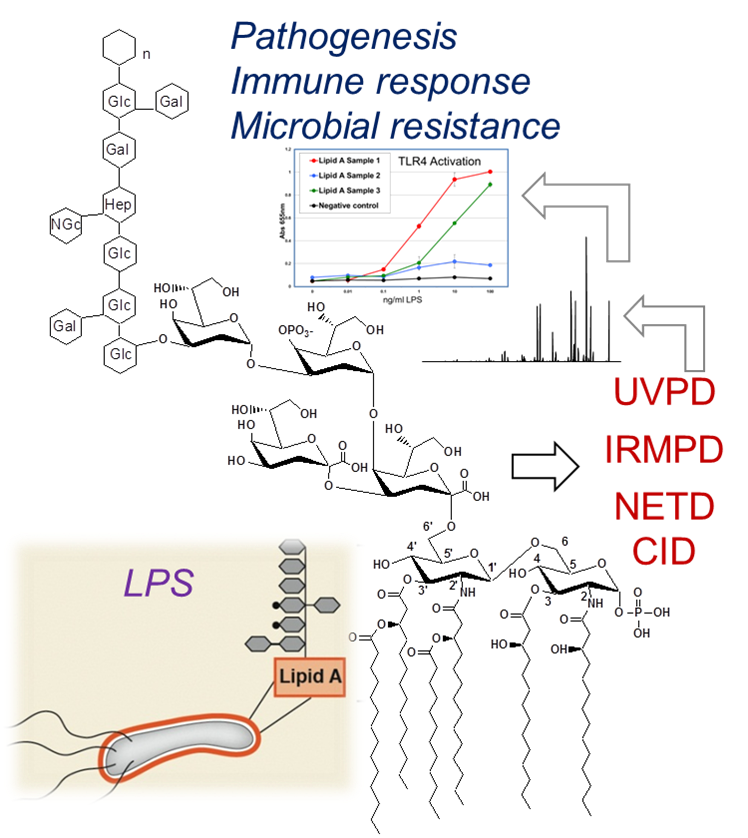
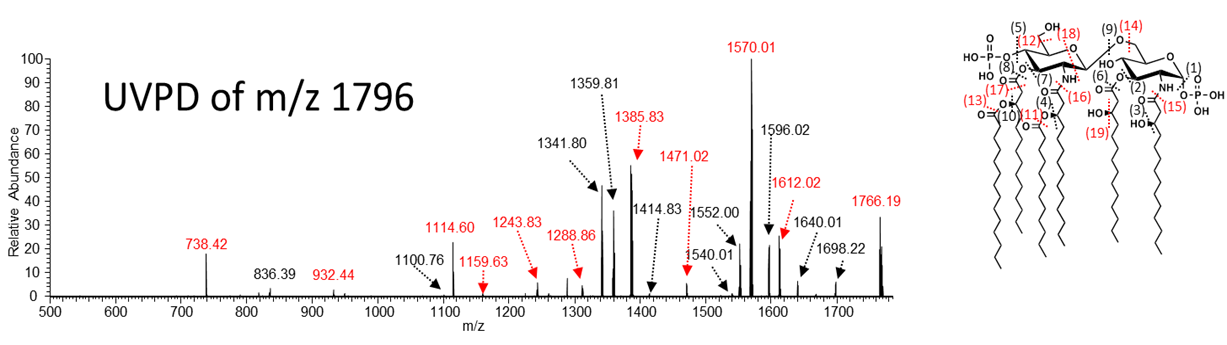
Structural Elucidation of Lipopolysaccharides
Gram-negative bacteria are responsible for some of the deadliest and more widespread pandemics in the world. Helicobacter pylori is now recognized as the primary cause of peptic ulcer disease and gastric cancer. Acinetobacter baumannii is responsible for a growing number of the antibiotic-resistant infections in hospitals. Understanding the machinery of these bacteria used for initiation of pathogenesis, for recognition and activation of the immune system, and for development of antibiotic resistance are vital issues that require multi-disciplinary research strategies. We are developing three photodissociation methods, including infrared multiphoton dissociation (IRMPD), ultraviolet photodissociation (UVPD), and activated–electron photodetachment dissociation, for the characterization of lipid A and LPS structures. Specific objectives include:
(1) Development of photodissociation and hybrid MS/MS methods for characterization of lipid A and core oligosaccharide/O-antigens.
(2) Applications to H. pylori, C. jejuni, V. cholera, and E. coli. In collaboration with Stephen Trent’s group, we are elucidating the LPS modification systems of these four proteobacteria.
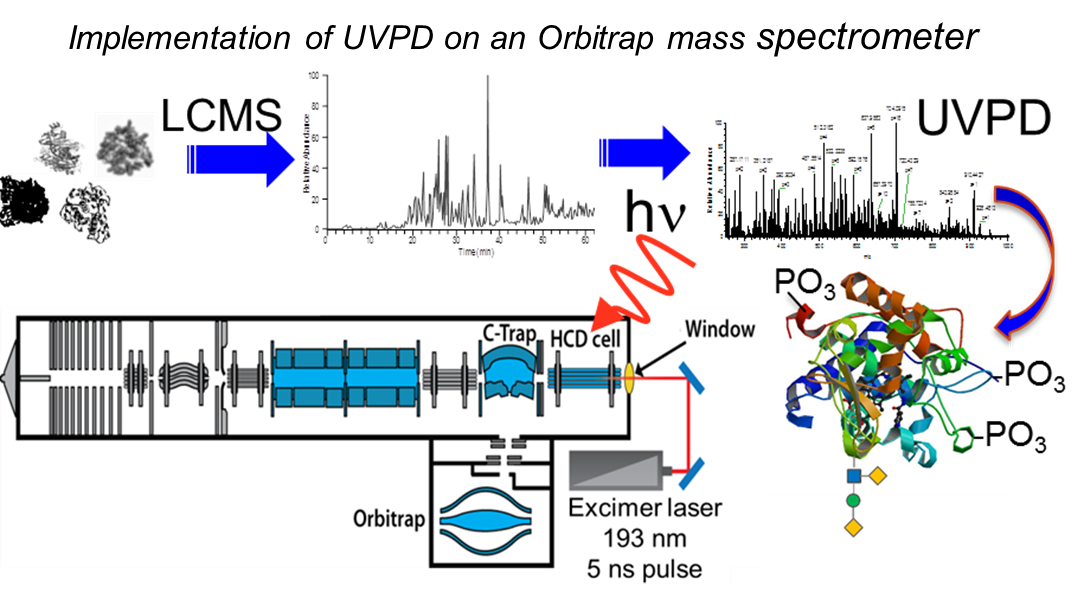
Top-Down Analysis of Intact Proteins
Recent advances in instrumentation and experimental design have propelled mass spectrometry to the forefront of proteome research. The top-down approach involves the interrogation of intact proteins, providing both intact protein and fragment mass measurements. UVPD affords broader sequence coverage than obtained using any other MS/MS method, and ion activation/dissociation can be accomplished using a single 5 nanosecond laser pulse. This translates to an MS/MS technology that can characterize intact proteins in exquisite detail, including mapping post-translational modifications that play critical roles in cell signaling and other functions. We are evaluating the analytical metrics of UVPD in an Orbitrap mass spectrometer relative to other MS/MS strategies with respect to sensitivity, speed and throughput, upper limit on molecular weight of protein, charge-state dependence, and sequence coverage.