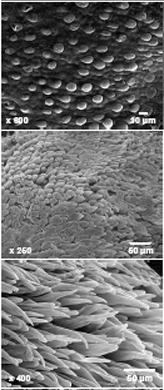
Cotton seed hair development share many similarities with Arabidopsis leaf trichome development [8], which is mediated by a “trichome activation complex”. Leaf trichome initiation in Arabidopsis thalianais promoted by the positive transcription regulators GLABROUS1 (GL1), TRANSPARENT TESTA GLABRA1 (TTG1), GLABRA3 (GL3), and ENHANCER of GL3 (EGL3) that are counteracted by the negative regulators TRIPTYCHON (TRY), CAPRICE (CPC), and ENHANCER of TRY and CPC1 (ETC1, 2, and 3) that encode single MYB-domain protein families [1, 4, 6, 12, 14]. GLABROUS2 (GL2) functions downstream of the GL1/TTG/GL3 complex and also plays a role in leaf trichome development [4, 13]. Some MYB factors such as AtMYB5 and AtMYB23 have minor effects on trichome initiation but regulate mucilage biosynthesis and seed coat development [9]. Moreover, trichome genes such as TTG1 and GL2 affect mucilage biosynthesis and columella cell formation [17, 19], suggesting a role of these trichome genes in seed coat development [3].
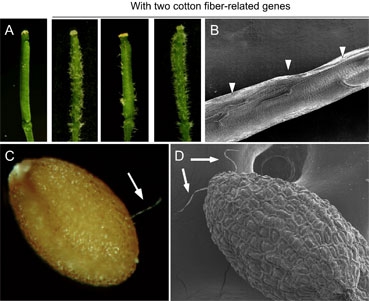
Figure 3. Production of ectopic trichomes and seed hair in Arabidopsis plants overexpressing two cotton fiber-related genes. (A) Siliques of Arabidopsis plants overexpressing a plasmid vector (left) or two cotton genes. (B) A scanning electron micrograph (SEM) showing ectopic trichomes produced inside of anArabidopsis silique. (C). Transgenic seed with a single hair. (D) A SEM image showing two seed hairs on a seed.
Several studies using cotton fiber-related genes have demonstrated a close relationship between cotton seed fibers and Arabidopsis leaf trichomes. Gossypium arboreum MYB2 (GaMYB2) encoding a putative homolog of GL1 MYB transcription factor complements the trichomeless gl1 mutant and induces occasional hair formation in A. thaliana seeds [16]. GaHOX1, a homeobox gene, encodes a HD-ZIP IV transcription factor, and is a functional homologue of the A. thaliana GL2 gene [2]. Two WD-repeat genes from Gossypium hirsutum (GhTTG1) restore trichome formation in A. thaliana ttg1mutant plants and complement anthocyanin defects in Matthiola incana ttg1 mutants [5]. Moreover, microrarray and gene expression analyses have uncovered many cotton fiber-related genes, including those encoding MYB transcription factors and phytohormonal regulators [7, 18]. For example, differential expression of six MYB genes is observed in allotetraploid cotton (G. hirsutum L.) [10]. Several MYB and RDL genes are expressed in fiber initials through microarray analysis [15]. GhMYB25 regulates early fiber and trichome development in cotton [11]. The data collectively suggest that Arabidopsis and cotton use similar transcription factors for the development of leaf trichomes and seed hairs. However, the mechanisms responsible for the differentiation of branched trichomes in vegetative tissues (leaves) and unbranched hairs in reproductive organs (seeds) may not be the same, and many seed plants including Arabidopsis do not produce seed hairs.
References
- Esch JJ, Chen MA, Hillestad M, Marks MD: Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. Plant J 40: 860-9 (2004).
- Guan XY, Li QJ, Shan CM, Wang S, Mao YB, Wang LJ, Chen XY: The HD-Zip IV gene GaHOX1 from cotton is a functional homologue of the Arabidopsis GLABRA2. Physiol Plant 134: 174-82 (2008).
- Haughn G, Chaudhury A: Genetic analysis of seed coat development in Arabidopsis. Trends Plant Sci 10: 472-7 (2005).
- Hülskamp M: Plant trichomes: a model for cell differentiation. Nat Rev Mol Cell Biol 5: 471-80 (2004).
- Humphries JA, Walker AR, Timmis JN, Orford SJ: Two WD-repeat genes from cotton are functional homologues of the Arabidopsis thaliana TRANSPARENT TESTA GLABRA1 (TTG1) gene. Plant Mol Biol 57: 67-81 (2005).
- Ishida T, Kurata T, Okada K, Wada T: A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59: 365-86 (2008).
- Lee JJ, Hassan OS, Gao W, Wei NE, Kohel RJ, Chen XY, Payton P, Sze SH, Stelly DM, Chen ZJ: Developmental and gene expression analyses of a cotton naked seed mutant. Planta: 1-15 (2005).
- Lee JJ, Woodward AW, Chen ZJ: Gene expression changes and early events in cotton fibre development. Ann Bot (Lond) 100: 1391-401 (2007).
- Li SF, Milliken ON, Pham H, Seyit R, Napoli R, Preston J, Koltunow AM, Parish RW: The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21: 72-89 (2009).
- Loguercio LL, Zhang JQ, Wilkins TA: Differential regulation of six novel MYB-domian genes defines two distinct expression patterns in allotetraploid cotton (Gossypium hirsutum L.). Mol Gen Genet 261: 660-671 (1999)
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES: The MYB transcription factor GhMYB25 regulates early fibre and trichome development. Plant J 59: 52-62 (2009).
- Pesch M, Hulskamp M: One, two, three...models for trichome patterning in Arabidopsis? Curr Opin Plant Biol 12: 587-92 (2009).
- Rerie WG, Feldmann KA, Marks MD: The GLABRA2 gene encodes a homeo domain protein required for normal trichome development in Arabidopsis. Genes Dev 8: 1388-99 (1994).
- Szymanski DB, Lloyd AM, Marks MD: Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci 5: 214-9. (2000).
- Taliercio EW, Boykin D: Analysis of gene expression in cotton fiber initials. BMC Plant Biol 7: 22 (2007).
- Wang S, Wang JW, Yu N, Li CH, Luo B, Gou JY, Wang LJ, Chen XY: Control of plant trichome development by a cotton fiber MYB gene. Plant Cell 16: 2323-34 (2004).
- Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW: MUCILAGE-MODIFIED4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134: 296-306 (2004).
- Wu Y, Machado AC, White RG, Llewellyn DJ, Dennis ES: Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 47: 107-27 (2006).
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A: A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859-69 (2003).