Cotton fibers are unicellular, unbranched, simple trichomes (or seed hairs) (Figure 2) differentiate from ~25% of the epidermal cells in the outer integument of a developing seed [2, 8, 14]. Visible signs of fiber development are first evident on the day of flower opening (anthesis) (Figure 2A), and subsequent development is described temporally by days post-anthesis (DPA). The fiber cells (Figure 2B) elongate to 3 to ~6 cm in G. hirsutum and G. barbadense, respectively, placing them among the largest plant cells. Elongation occurs via synthesis of a thin (0.1 – 0.2 µm) primary wall with a composition typical of many plant cells [9]. Facilitated by the cell wall-mediated coordination of the packing of elongating fibers [12], half to 3/4 million fibers differentiate on 24-30 seeds within one cotton fruit (or boll) (Figure 2C). The percentage of cellulose in cotton fibers is far higher than most plant cells partly because the thick secondary wall in the fibers consists of nearly pure cellulose [8]. G. barbadense produces longer, stronger, and finer fiber than G. hirsutum. (Finer means less mass per length, which can arise from a thinner cell wall and/or a smaller diameter fiber.) Typically G. barbadense has a longer phase of fiber elongation compared to G. hirsutum [6, 11], but it is unknown how elongation is sustained in G. barbadense even though secondary wall deposition has begun. The differences in fiber traits may reflect convergent evolution of useful fiber morphology with a different genetic basis. For example, A-genome ESTs are enriched during early stages of fiber development [15], but the D-genome expression was enhanced during domestication, and different sets of genes showed this bias in the two separate domestication events [7]. The ability to introduce key fiber quality genes from G. barbadense into agronomically more robust G. hirsutum depends on identifying all key cellular differences and regulatory genes. Several groups have compared the fiber transcriptomes and/or analyzed recombinant inbred lines in the two species [1, 10, 13], as well as identified the QTLs related to fiber elongation, fineness, and length using a backcross population between G. hirsutum L. and G. barbadense L. [3-5].
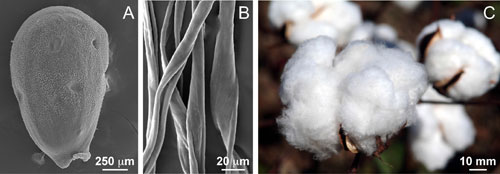
Figure 2.Fiber cell initiation from epidermal layer of an ovule on the day of anthesis (0 DPA) (A), elongated cotton fibers under electron microscope (B), and mature cotton bolls (C).
References
- Al-Ghazi Y, Bourot S, Arioli T, Dennis ES, Llewellyn DJ: Transcript profiling during fiber development identifies pathways in secondary metabolism and cell wall structure that may contribute to cotton fiber quality. Plant Cell Physiol 50: 1364-81 (2009).
- Basra A, Malik CP: Development of the cotton fiber. Int Rev Cytol 89: 65-113 (1984).
- Chee P, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor Appl Genet 111: 757-63 (2005).
- Chee PW, Draye X, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of phenotypic variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: III. Fiber length. Theor Appl Genet 111: 772-81 (2005).
- Draye X, Chee P, Jiang CX, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH: Molecular dissection of interspecific variation between Gossypium hirsutum and G. barbadense(cotton) by a backcross-self approach: II. Fiber fineness. Theor Appl Genet 111: 764-71 (2005).
- Hawkins RS, Serviss GH: Development of cotton fibers in the Pima and Acala varieties. J. Agriculture Research 40: 1017-1029 (1930).
- Hovav R, Chaudhary B, Udall JA, Flagel L, Wendel JF: Parallel domestication, convergent evolution and duplicated gene recruitment in allopolyploid cotton. Genetics 179: 1725-33 (2008).
- Kim HJ, Triplett BA: Cotton fiber growth in planta and in vitro: Models for plant cell elongation and cell wall biogenesis. Plant Physiol 127: 1361-6 (2001).
- Meinert MC, Delmer DP: Changes in Biochemical Composition of the Cell Wall of the Cotton Fiber During Development. Plant Physiol 59: 1088-1097 (1977).
- Ruan YL, Xu SM, White R, Furbank RT: Genotypic and developmental evidence for the role of plasmodesmatal regulation in cotton fiber elongation mediated by callose turnover. Plant Physiol 136: 4104-13 (2004).
- Schubert AM, Benedict CR, Gates CE, Kohel RJ: Growth and development of the lint fibers of Pima S-4 cotton. Crop Sci 16: 539-543 (1976).
- Singh B, Avci U, Eichler Inwood SE, Grimson MJ, Landgraf J, Mohnen D, Sorensen I, Wilkerson CG, Willats WG, Haigler CH: A specialized outer layer of the primary cell wall joins elongating cotton fibers into tissue-like bundles. Plant Physiol 150: 684-99 (2009).
- Tu LL, Zhang XL, Liang SG, Liu DQ, Zhu LF, Zeng FC, Nie YC, Guo XP, Deng FL, Tan JF, et al.: Genes expression analyses of sea-island cotton (Gossypium barbadense L.) during fiber development. Plant Cell Rep 26: 1309-20 (2007).
- Wilkins TA, Jernstedt JA: Chapter 9. Molecular genetics of developing cotton fibers. In: Basra AM (ed) Cotton Fibers, pp. 231-267. Hawthorne Press, New York (1999).
- Yang SS, Cheung F, Lee JJ, Ha M, Wei NE, Sze SH, Stelly DM, Thaxton P, Triplett B, Town CD, et al.: Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47: 761-775 (2006).