Swarming, Motor Function, Flagellar Biogenesis, Necrosignaling, c-di-GMP
|
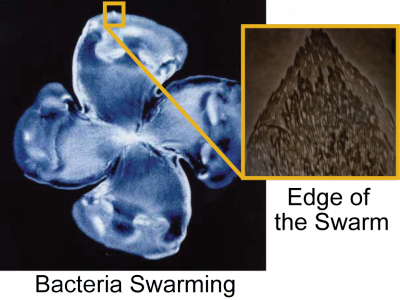
Swarming |
|
|
|
Swimming bacteria use chemotaxis to find nutrients and avoid toxic environments, while swarming bacteria appear to suppress chemotaxis and to use the dynamics of their collective motion to expand and acquire new territory, barrel through lethal chemicals, carry along bacterial and fungal cargo, and engage in group warfare for niche dominance. Currently, our lab focuses on two aspects of swarming: (1) how bacteria sense they are on a surface and turn on programs that promote movement, and (2) how do swarming bacteria override scarcity and adversity as dense packs.
|
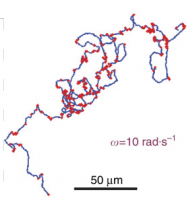
| Bacterial Trajectories | |
|
We have fluorescently labeled only a few bacteria moving within a swirling, billowing, swarming mass (discernible in the grey background), so we can track their individual motion. We discovered that bacteria are doing a dance called the Levy Walk, characterized by sudden long jumps. This migration pattern has been observed in times of scarcity in large animals, including humans. It is thought that the long forays help maximize search of random areas. We are excited at this discovery, which opens up a whole new area of investigation. For a visual snippet of this, Click Here. |
|
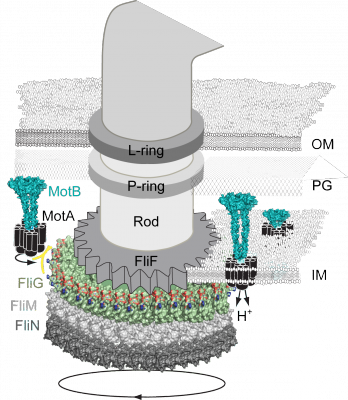
| Motor Function | |
|
I. Torque Generation Bacterial flagella rotate up to 1700 Hz, five times faster than a formula-one racecar engine. In almost every motile bacterial species, integral transmembrane protein complexes (stators, MotA4B2) initiate and maintain flagellar rotation. Interactions between MotA and FliG are necessary for rotation; however the mechanism of torque generation is unknown. We seek to answer the long standing question, how is torque generated for flagellar rotation? II. The Motor as a Sensor YcgR is a c-di-GMP receptor that interacts with the flagellar motor to skew the bias to CCW rotation ultimately decreasing motor speed. It is known that YcgR interacts with FliM, FliG and MotA; however, it still remains to be understood how exactly YcgR restricts motor rotation. |
|
|
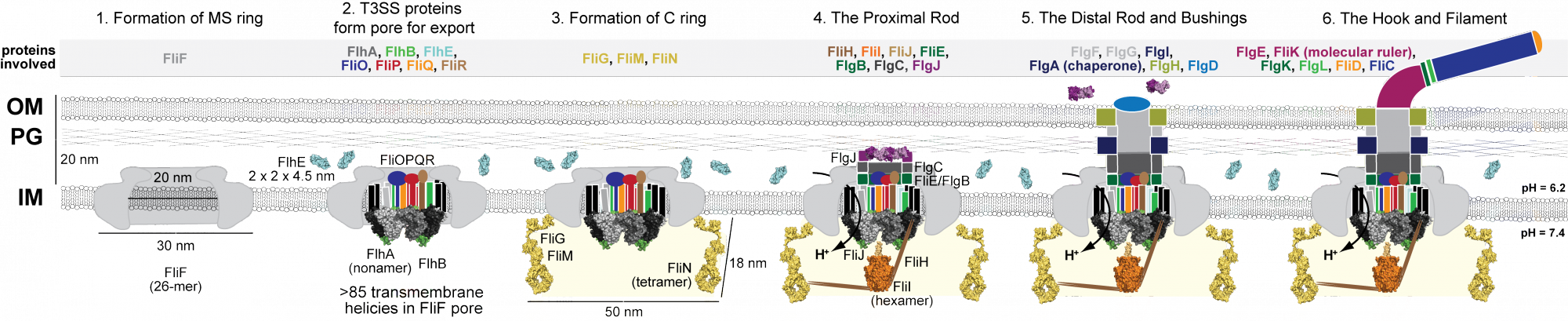
Flagellar Biogenesis |
|
|
Over 40 proteins symphonically assemble to build a single bacterial flagellum. The structure and function of many, but not all, flagellar proteins is known. We are currently targeting FlhE, FliL and FliF for structure-function studies. |
|
|
|
Necrosignaling
Many bacterial swarms display a temporary, non-mutational, and reversible resistance to antibiotics in a phenomenon called as ‘adaptive resistance. We discovered the process of ‘necrosignaling’ where dead cells of an E. coli swarm population release a periplasmic protein AcrA that binds to the outer membrane protein TolC of live cells within the same population. This binding acts as a signal to increase the activity of efflux pumps which contributes to the establishment of antibiotic resistance. Investigating whether other bacteria such as Bacillus and Pseudomonas also possess this mechanism will shed light on the universality of this phenomenon.
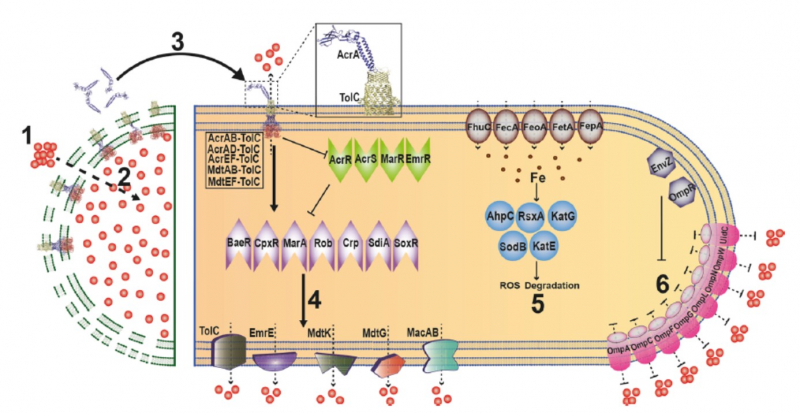
1. Antibiotic uptake (red dots). 2 Cell death, membrane damage. 3 Released AcrA binds to TolC in the OM of live cells, activating efflux from TolC pumps. 4 A secondary consequence of TolC activation is upregulation of five categories of efflux pumps. This upregulation is enabled by mulitple activators (purple), or by repression of multiple repressors (green). 5 Upregulation (arrowhead) of genes that reduce ROS (blue and brown) would allow tolerance of the antibiotic stress. 6 Downregulation (flathead) of porins (purple) would restrict entry of antibiotics. (Figure from Bhattacharyya et al., Nat Comm., 2020)
|
c-di-GMP |
|
|
|
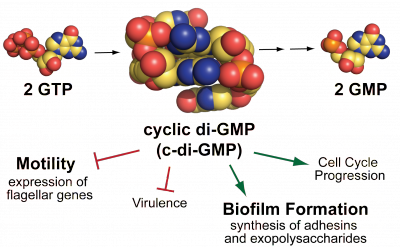
The cyclic dinucleotide c-di-GMP is a signaling molecule that mainly controls the transition between motility and biofilm formation. While seeking c-di-GMP signaling proteins involved with motility, we found that the c-di-GMP synthetic enzyme YfiN localizes to the division site in Salmonella and E. coli. Investigating how YfiN regulates both c-di-GMP levels and cell division will provide insight into he versatility of c-di-GMP signaling proteins in bacteria. |