DNA Transposition, DNA Repair, Genome Organization
We are interested in the biology of bacteriophage Mu, which is unique among the phage world for its ability to replicate as a transposon. Mu is a lysogenic phage, capable of inserting into the host chromosome like a transposon. In the lytic cycle, Mu replicates by transposing randomly across the host chromosome. The transposition of Mu during the lytic cycle is estimated to occur about 100 times, revealing a robust transposition mechanism. The chemical nature of Mu’s transposition reaction is known (Fig. 1). While DNA cleavages are carried out the transposase MuA, a target capture protein MuB is responsible for the efficiency of Mu transposition. A partial structure of the transpososome assembled on 4 of 6 binding sites for the transposase at the two ends of Mu is available. However, transpososome assembly requires engagement of the two ends with a third enhancer site. The structure of this 3-site complex is not yet determined. The size of phage Mu is about 37kb, about a hundred times smaller the E. coli’s genome. Given the random nature of the transposition reaction, Mu can be expected to insert within itself. Yet, that does not happen. This phenomenon is termed Mu-immunity and is different from the cis-immunity mechanism where regions outside and adjacent to Mu ends are also protected. The unique replication lifestyle of Mu poses multiple interesting questions:
- What is the structure of the transpososome that enables multiple rounds of transposition across the host chromosome?
- What mechanisms mediate Mu-immunity?
-
How can we employ the random nature of Mu insertion to probe the host chromosomal structure?
I. Mu Transposition Mechanism
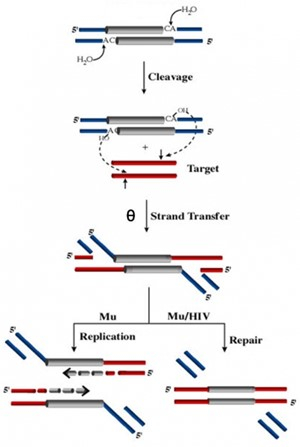
In the figure to the right (Figure 1), note the similarities between the Mu and HIV systems: Both genomes are flanked by the terminal dinucleotide CA which is cleaved to yield 3' OH groups. The strand transfer or joining step is identical in both systems and leads to integration of the viral genome into the host (target) genome to produce a θ intermediate. The chemistry of this class of phosphoryl transfer reactions is common to immunoglobulin rearrangements and RNA splicing. Unlike known transposable elements, which move by some one particular mechanism, Mu transposes by two alternative pathways - replicative or non-replicative transposition - depending on the phase of its life cycle. Both pathways go through a common θ intermediate but process this intermediate differently. During integration of infecting Mu, the θ intermediate is repaired without replication, while during lytic growth it is resolved by replication. In the non-replicative repair pathway, the flanking DNA or 5' flaps attached to the ends of Mu are removed. We have discovered that the MuA transposase itself is responsible for flap removal. The flap endonuclease activity is encoded in the C-terminal domain of MuA, distinct from the DDE domain that carries out the cleavage and strand transfer steps of transposition. This activity is normally masked in the full-length protein and has only been observed in vivo. Flap removal requires the host ClpX protein, which is known to interact with the C-terminus of MuA to remodel the transpososome for replication. We hypothesize that ClpX constitutes part of a highly regulated mechanism that unmasks the cryptic endonuclease activity of MuA specifically in the repair pathway. We have begun to actively study this pathway, which is also used by HIV, and is currently completely unexplored. In addition to ClpX, we identified 5 proteins involved in double-strand break (DSB) repair that are essential during the non-replicative pathway. We discovered that upon integration of infecting Mu, the assembled transpososome waits for the E. coli replisome, arrival of which results in a DSB. The communication between and the replisome that initiates repair of the transposition intermediate is a fascinating problem to solve.
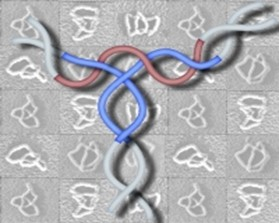
We have also examined the topology of the transpososome. The Mu transpososome is assembled by interactions of transposase MuA subunits with the left and right ends of Mu and an enhancer located in between. We have determined the path of the DNA within the three-site LER synapse using a method we call 'difference topology'. As shown to the right (Figure 2), the three sites (enhancer is red and the two Mu ends are blue) interwrap around each other five times. This is the most complex DNA arrangement seen to date within a recombination synapse. Solving it has posed a challenge thus far because this structure can only be assembled on supercoiled DNA.
Transposition is a double-edged sword, allowing elements t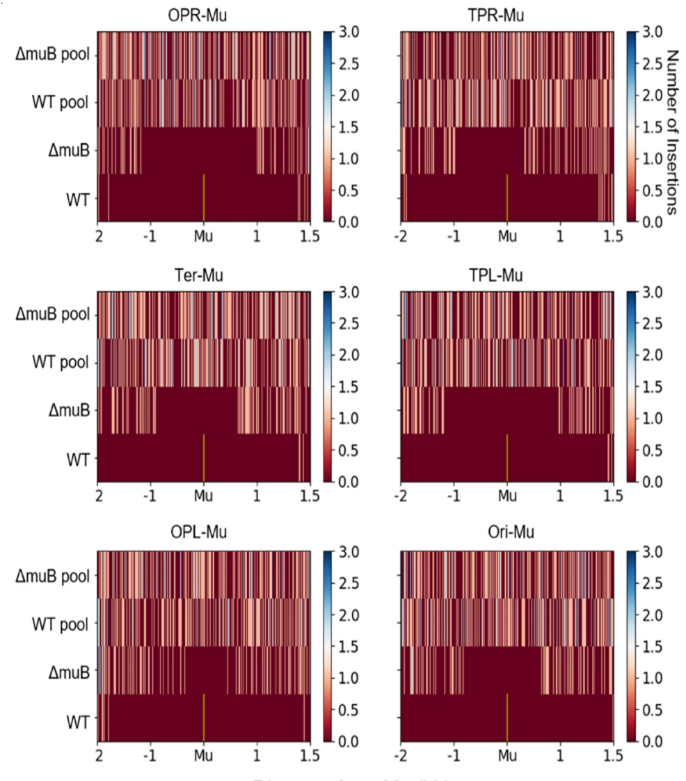
II. Mu as a Probe for Genome Structure
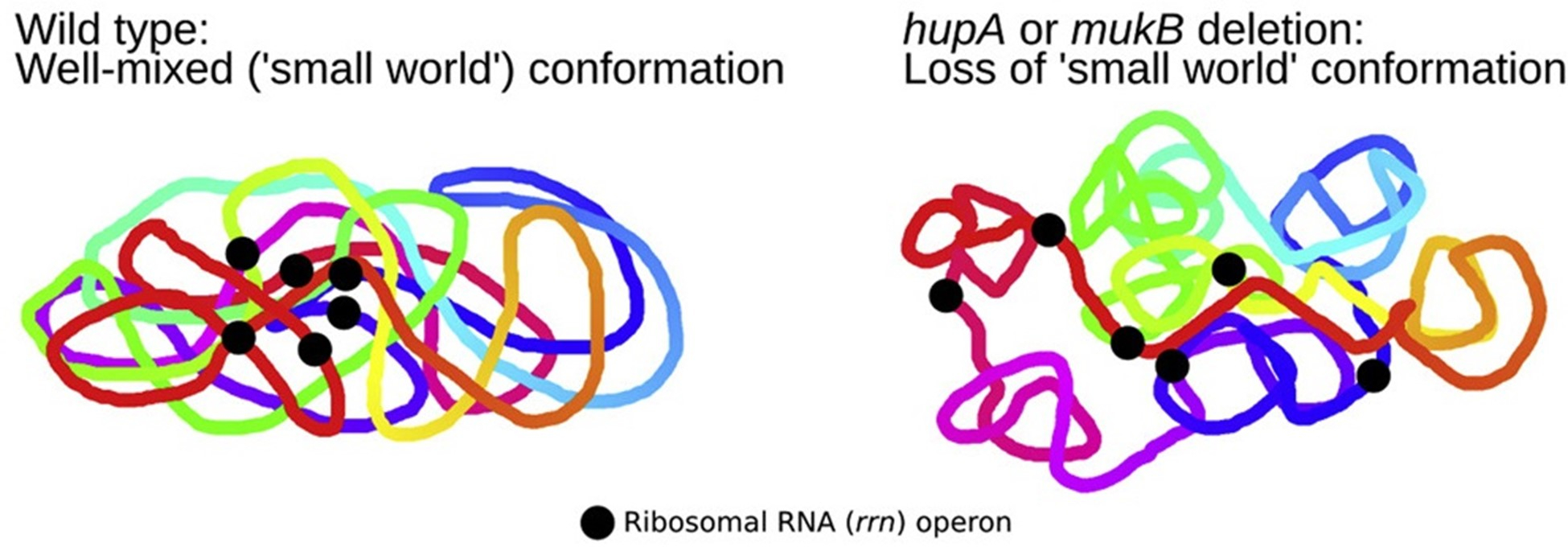
The random nature of Mu insertion suggest that Mu insertion could be used as a probe for long-range interactions in the host chromosome. We recently developed a method based on transposition of the phage Mu to identify long-range contacts in the E. coli chromosome. 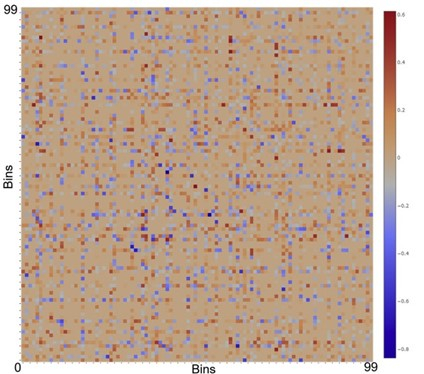 The Mu-based methodology requires no chemical fixation or external perturbation. Rather than observing static snapshots of a chemically fixed genome, Mu captures the live dynamic nature of the genome, and provides unambiguous data in the form of new chromosomal integrations of the Mu sequence that can be read by whole genome sequencing or amplicon sequencing. Unlike the dominance of short-range contacts seen with crosslinking-based methodologies, Mu reports the formation of many long-range contacts that we subsequently verified via fluorescence microscopy (Figure 4). The contacts between functionally related genes, or ‘cluster’, suggesting a mode of gene regulation. In addition, we also identified multiple proteins that are responsible for these clustering events. Collectively, our data suggest that long-range DNA contacts in the E. coli’s chromosome is much more frequent than previously thought, thus providing a novel view of the E. coli’s chromosome (Figure 5). We are currently working on understanding the molecular nature of the clusters as well as expanding the Mu methodology to other bacteria and eukaryotes.
The Mu-based methodology requires no chemical fixation or external perturbation. Rather than observing static snapshots of a chemically fixed genome, Mu captures the live dynamic nature of the genome, and provides unambiguous data in the form of new chromosomal integrations of the Mu sequence that can be read by whole genome sequencing or amplicon sequencing. Unlike the dominance of short-range contacts seen with crosslinking-based methodologies, Mu reports the formation of many long-range contacts that we subsequently verified via fluorescence microscopy (Figure 4). The contacts between functionally related genes, or ‘cluster’, suggesting a mode of gene regulation. In addition, we also identified multiple proteins that are responsible for these clustering events. Collectively, our data suggest that long-range DNA contacts in the E. coli’s chromosome is much more frequent than previously thought, thus providing a novel view of the E. coli’s chromosome (Figure 5). We are currently working on understanding the molecular nature of the clusters as well as expanding the Mu methodology to other bacteria and eukaryotes.