The ability of TGIRT-seq to comprehensively profile essentially all RNA biotypes from small amounts of starting material makes it useful for RNA-based diagnostics, including the analysis of cell-free RNAs (cfRNAs), which are present in low concentrations in human plasma and other bodily fluids. cfRNAs are secreted in extracellular vesicles (EVs), such as exosomes, or released from cells undergoing apoptosis or necrosis, and they have been of wide interest because of their potential involvement in intercellular communication and as potential biomarkers for human diseases. Recent studies in our laboratory demonstrated the utility of TGIRT-seq for the analysis of human EV and plasma RNAs and suggest new approaches for liquid biopsy and RNA diagnostics. These studies have also drawn our attention to 3’-tRNA fragments (3’-tRFs), an emerging class of small regulatory RNAs. TGIRTs are now being used for the liquid biopsy component in a Phase III clinical trial of the efficacy of an advanced breast cancer drug in collaboration with physicians at M.D. Anderson
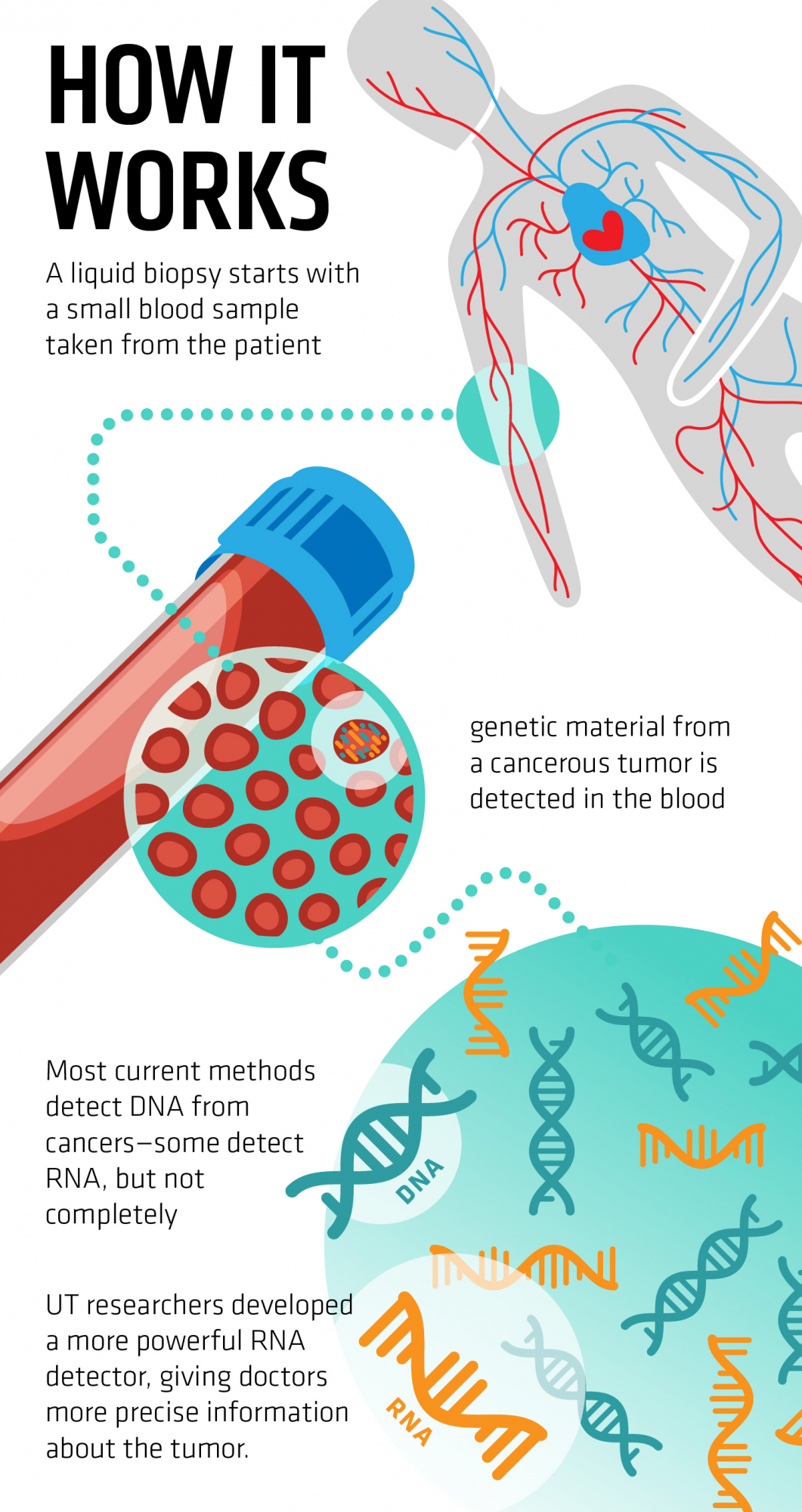
Image source: https://news.utexas.edu/2017/11/16/improving-cancer-detection-with-liqui...
Selected Publications:
Yao J, Wu DC, Nottingham RM, Lambowitz AM. Identification of protein-protected mRNA fragments and structured excised intron RNAs in human plasma by TGIRT-seq peak calling. eLife [Internet]. 2020.
Reinsborough, C.W.*, Ipas, H.*, Abell, N.S.*, Nottingham, R.M.*, Yao, J., Devanathan, S.K., Shelton, S.B., Lambowitz, A.M. and Xhemalce, B. BCDIN3D regulates tRNAHis 3’ fragment processing. PLoS Genetics. 2019;15 (7).
Temoche-Diaz, M.M., Shurtleff, M.J., Nottingham, R.M., Yao, J., Fadadu, R.P., Lambowitz, A.M., and Schekman, R. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. bioRxiv, doi.org/10.1101/612069, 2019.
Wu, D.C. and Lambowitz, A.M. Facile single-stranded DNA sequencing of human plasma DNA via thermostable group II intron reverse transcriptase template switching. Scientific Reports, 7:8412, DOI:10.1038/s41598-017-09064-w, 2017.
Shurtleff, M.J., Yao, J., Qin, Y., Nottingham, R.M., Temoche-Diaz, M., Schekman, R., and Lambowitz, A.M. A broad role for YBX1 in defining the small non-coding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA, 114(43):E8987-E8995, 2017.
Carrell, S.T., Tang, Z., Mohr, S., Lambowitz, A.M., and Thornton, C.A. Detection of expanded RNA repeats using thermostable group II intron reverse transcriptase. Nucleic Acids Res., 46(1):e1. doi: 10.1093/nar/gkx867, 2017.
Qin, Y., Yao, J., Wu, D.C., Nottingham, R., Mohr, S., Hunicke-Smith, S., and Lambowitz, A.M. High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22, 111-128, 2016.


