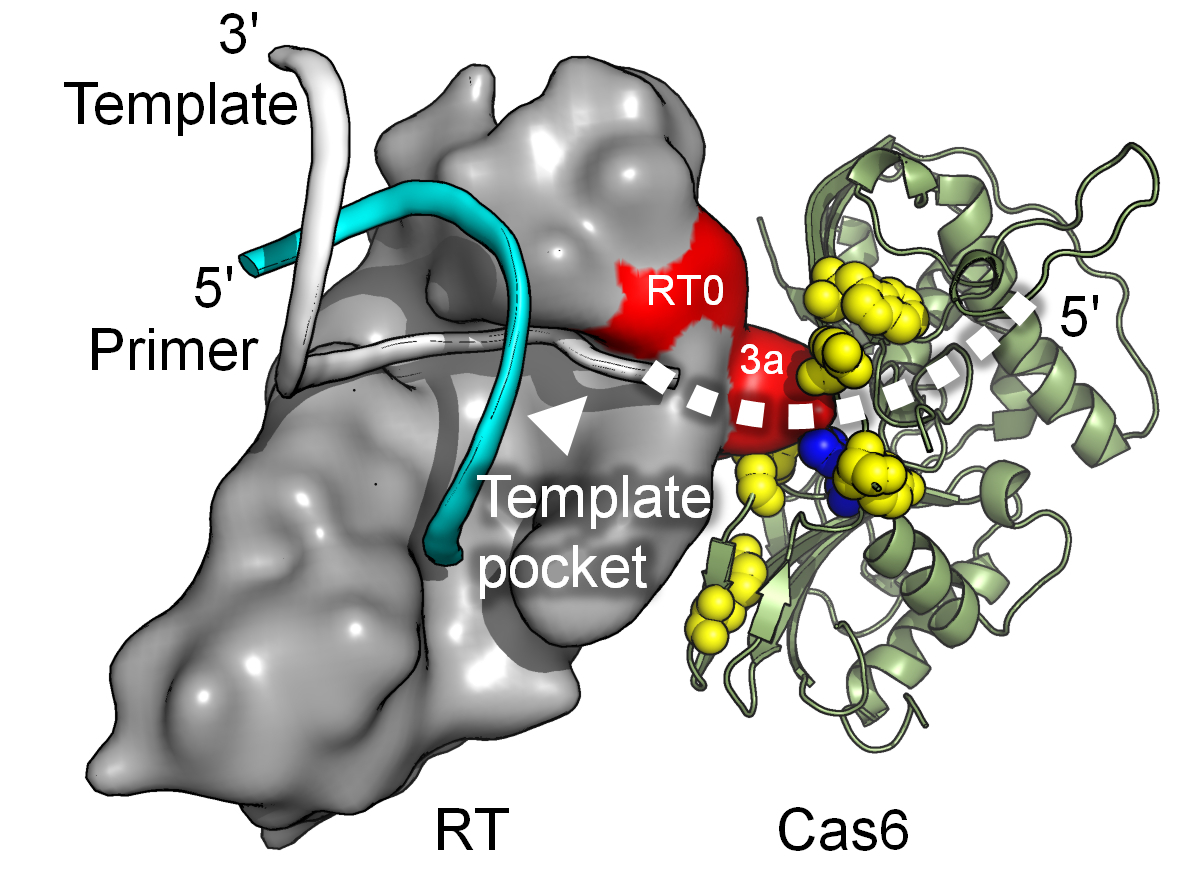
In addition to group II intron RTs, bacteria contain a variety of other closely related RTs, some of which have evolved cellular functions. These include retron RTs; diversity-generating retroelement RTs, which function in host-phage tropism switching; abortive phage infection RTs, which function in phage defense by unknown mechanisms; RTs associated with CRISPR systems; and a variety of putative RTs of unknown function encoded by conserved ORFs in bacterial genomes. We are interested in these RTs because of their potential to provide new insights into the biological functions and biochemical mechanisms of RTs, as well as new RTs with desirable properties for biotechnological applications. In collaboration with Dr. Andrew Fire (Stanford), we identified an active RT-Cas1 fusion protein associated with a Type III CRISPR system and showed that it functions to site-specifically integrate RNA spacers into CRISPR arrays by a novel mechanism with features resembling group II intron retrohoming. Going forward, we will continue to study CRISPR-associated RTs and extend this research to other families of bacterial RTs. We are also interested in returning to a novel mitochondrial retroplasmid RT that functions like an RNA-depended RNA polymerase in recognizing a 3’ tRNA-like structure of the plasmid transcript and has the unprecedent ability to initiate cDNA de novo (i.e., without a primer).
Selected Publications:
Mohr, G., Silas, S., Stamos, J.L., Makarova, K.S., Markham, L.M., Yao, J., Lucas-Elio, P. Sanchez-Amat, A., Fire, A.Z., Koonin, E.V., and Lambowitz, A.M. A Reverse Transcriptase-Cas1 Fusion Protein Contains a Cas6 Domain Required for Both CRISPR RNA Biogenesis and RNA Spacer Acquisition. Mol. Cell, 72, 700-714, 2018.

