Retroviral RTs, which are currently used for RNA-seq and other biotechnological applications, have inherently low fidelity and processivity to help retroviruses evade host defenses, and the extent to which these properties can be improved by protein engineering or in vitro evolution is limited by the retroviral RT structural framework. By utilizing the beneficial properties of thermostable group II intron RTs (TGIRTs), we developed TGIRT-based RNA-seq methods (TGIRT-seq) with advantages over current methods, including better RNA-seq metrics, the ability to obtain full-length end-to-end reads of tRNAs and other structured small ncRNAs (sncRNAs), and the ability to simultaneously profile mRNAs and lncRNAs together with sncRNAs and miRNAs in the same RNA-seq. As a result of these studies, TGIRTs are now sold commercially and have used for a variety of RNA-seq applications, including applications that would be difficult or impossible with retroviral RTs.
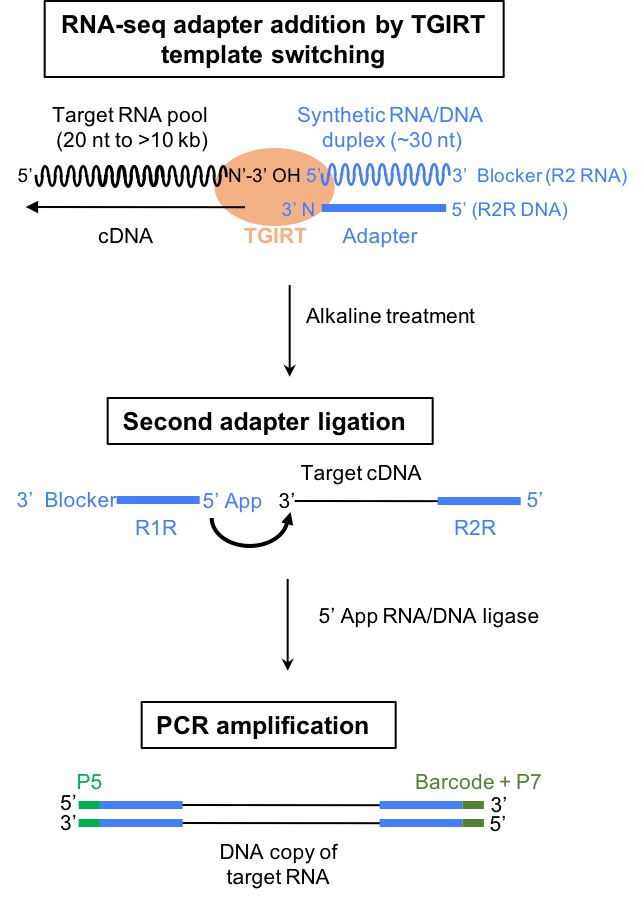
Selected Publications:
Yao J, Wu DC, Nottingham RM, Lambowitz AM. Identification of protein-protected mRNA fragments and structured excised intron RNAs in human plasma by TGIRT-seq peak calling. eLife [Internet]. 2020.
Xu, H., Yao, J., Wu, D.C., and Lambowitz, A.M. Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer-formation and bias correction. Scientific Reports, 9(1), 2019.
Boivin, V., Deschamps-Francoeur, G., Couture, S., Nottingham, R.M., Bouchard-Bourelle, P., Lambowitz, A.M., Scott, M.S., and Abou-Elela, S., Simultaneous sequencing of coding and non-coding RNAs reveals a human transcriptome dominated by a small number of highly expressed non-coding genes. RNA, 24, 950-965, 2018. doi:10.1261/rna.064493.117
Zubradt, M., Gupta, P., Persad, S., Lambowitz, A.M., Weissman, J.S., and Rouskin, S. DMS-MaPseq for genome-wide or targeted RNA structure probing in vivo. Nature Methods, 14, 75-82, 2017.
Nottingham, R.M., Wu, D.C., Qin, Y., Yao, J., Hunicke-Smith, S., and Lambowitz, A.M. RNA-seq of human reference RNA samples using a thermostable group II intron reverse transcriptase. RNA, 22, 597-613, 2016.
Zheng, G..#, Qin, Q.#, Clark, W.C., Yi, C., He, C., and Lambowitz, A.M.* and Pan, T.* Efficient and quantitative high-throughput transfer RNA sequencing. Nature Methods, 12, 835-837, 2015. #Co-first authors, *Co-corresponding authors. [Commentary Wilusz, J.E. Removing roadblocks to deep sequencing of modified RNAs. Nature Methods, 12, 831-832, 2015.]
Shen, P.S., Park, J., Qin, Y., Li, X., Parsawar, K., Larson, M., Cox, J., Cheng, Y., Lambowitz, A.M., Weissman, J.S., Brandman, O., and Frost A. Rqc2p and the 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75-78, 2015.
Katibah, G.E., Qin, Y., Sidote, D.J., Yao, J., Lambowitz, A.M. and Collins, K. Broad and adaptable RNA structure recognition by the human interferon-induced tetratricopeptide repeat protein IFIT5. Proc. Natl. Acad. Sci., USA, 111, 12025-12030, 2014.
Mohr, S., Ghanem, E., Smith, W., Sheeter, D., Qin, Y., King, O., Polioudakis, D., Iyer, V.R., Hunicke-Smith, S. Swamy, S., Kuersten, S., and Lambowitz, A.M. Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation sequencing. RNA 19, 958-970, 2013.
Commentary: Collins, K. and Nilsen, T. W. Enzyme engineering through evolution: Thermostable recombinant group II intron reverse transcriptases provide new tools for RNA research and biotechnology. RNA 19, 1089-1104, 2013.


